| HOME | | | TEAM | | | CONTACT US | | | DOCUMENTATION |
|
LeishMicrosatDB Documentation
About LeishMicrosatDBLeishMicrosatDB is a complete curated web-oriented relational database of perfect, compound and clusters of repeats along with their locus coordinates in eight sequenced trypanosomatids genome. It contains about 1,738,669 microsatellite repeats from six leishmania species of which we have predicted about 61,624 repeats are polymorphic. The repeats can be searched in a chromosome using following need based input parameters like repeat type (mono- hexa), coding status, Repeat unit length and repeat sequence motif. How to search Perfect Microsatellites?Each Chromosome of any species can be searched for Simple repeats. User needs to specify atleast a species name. Selecting species automatically loads chromosomes. To narrow-down search, user need to select required options. These options include specifying basic parameters like repeat type and coding status of the repeat. Search can be further narrowed down to specific repeat motif selecting Advanced search option. User needs to be careful when specifying Motif. If he has selected repeat type as Trinucleotide in basic search option, then it is required to enter a motif of length 3. To search a specific portion of the selected chromosome user may provide start and end coordinates in base pairs. To find repeats present within a single loci, Locus specific search may be performed. example:If we desire to search all SSR repeats with having repeating units greater than four in L. donovani. Select Leishmania donovani in Select species dropdown box. Set chromosomes to 'all'. Click on Advanced search tick mark. set repeating units greater than 4. click submit.
How to search Compound Microsaetellites?Those repeating units which have more than one type, one merged with another are called compound SSR. E.g (CA)8(CG)12. user can search compound repeats in a single chromosome or in all chromosmes. This search has been designed to search compond repeats of specific combination. So user needs to build a search query. For building a query user need to select a repeat type, if desired enter a repeat mootif of eqivalent length and repeating units; and click on add to query button. He can iterate this operation for creating a combination. User can specify a desired maximum length for the compound repeat. If user desires to search all the possible combination of repeats in te chromosome, simply enter 'all' in the query filed without quotes. example:if user wants to find Compound Microsaetellites in L. donovani chromosome 4. It requires to find all repeats with a combination of di-nucleotides having repeating units greater than 4 and it must be followed by a mono-nucleotide of reating units greater than 5. All the compound repeats must be at least 10 bp long. Select the species and chromosome number. Select repeat type di-nucleotide. repeating unit greater than 4. click Add to query button. This will add the first term of the compound repeat. Again repeat the same process for mono-nuceloteide of repeting unit 5. click add to query. And teh query is ready. Enter length as 10. Submit to search.
How to search Microsatellite clusters?Sometime microsatellites aggregate among themselves to form a cluster. User can identify such clusters by defining a maximum interruption distance between two adjacent repeats. Such cluster of desired maximum length can be defined and searched. example:For finding repeat clusters in L.donovani chromosome number 14 having maximum iterruption length of 10 bp, which are presnt in the genic reion and they are of atleast 20 bp long. Set all the parameters as shown in the snapshot and click submit.
How to find polymorphic Microsatellites?Microsatellite loci those can be PCR amplified wih identical primers but are having different repeating units are called as polymorphic microsatellite loci. e.g. (CA)4 and (CA)6. Using the existing SSR loci we have used comparative genomics approach to mine polymorphic microsatellite loci among these six leishmania species. In order to search polymorphic repeats user needs to select a reference chromosomes from any species. When a single species is seleced all other spcies are trated as another group with respect to which SSR loci are termed as polymorphic. This group contains a list of species names separated by commae(,) and can be edited to alter tha search space. User can remove name of the species from the comparing group and submit for polymorphic repeating finding in the reference genome with the desired comparing group. Select required repeat units and repeat types. User can specify the location in the genic or intrgenic region.
example:We need to find Polymorphic SSRs(PSSRs) in chromosme 6 of L. donovani. The rpeat type includes all the repats except mono-nucleotides and repeats must be having repeat unit greate than 4.
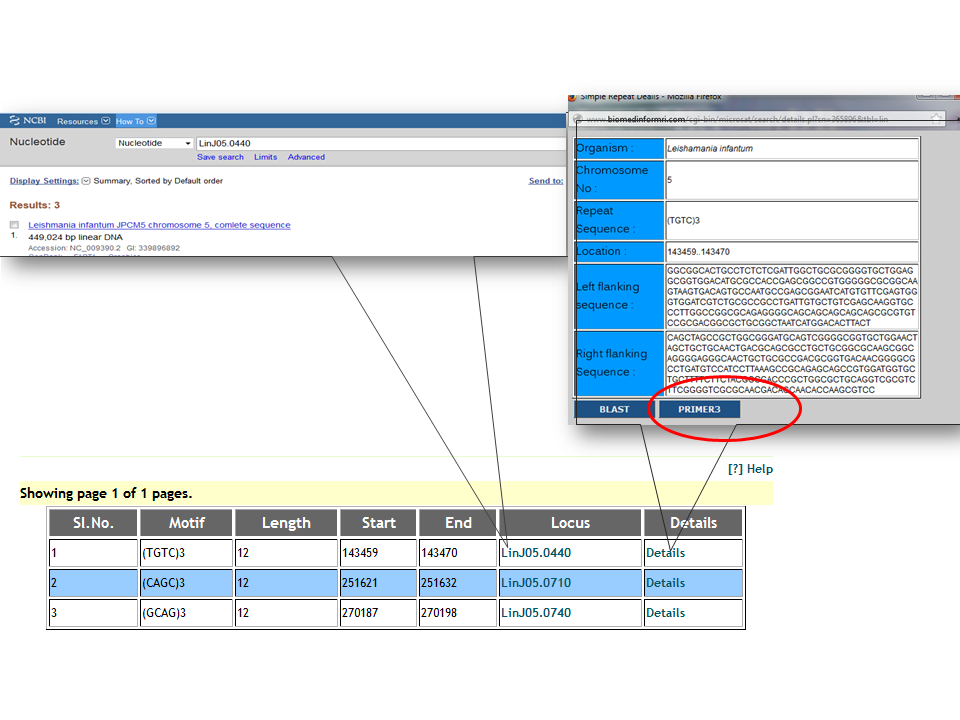
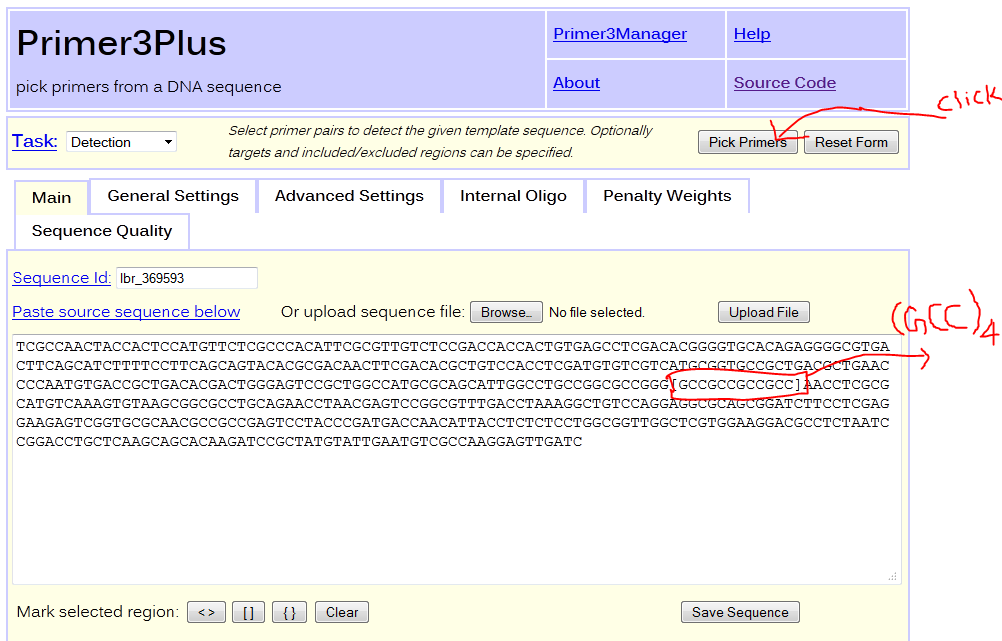
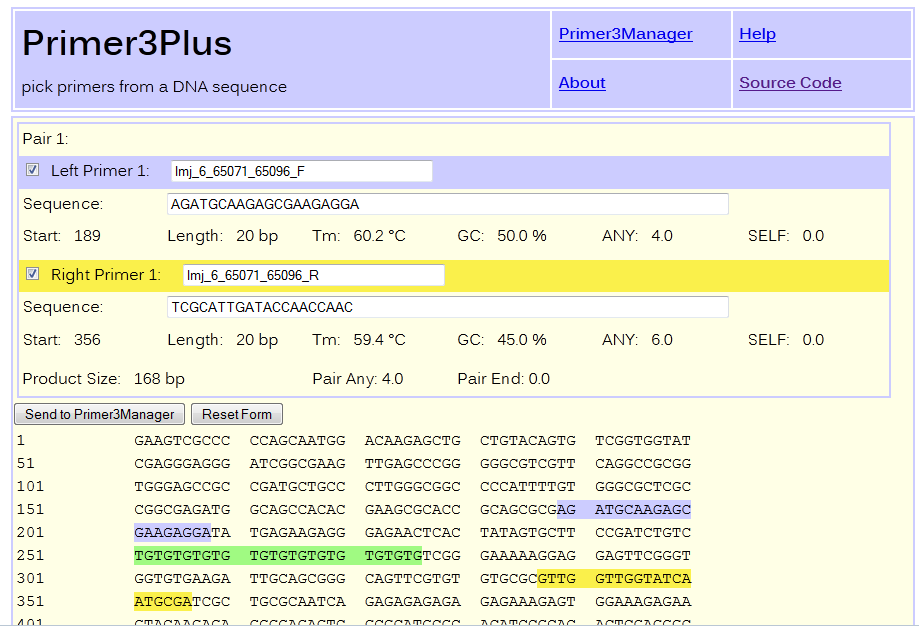
How to design a PCR primer for a selected locus?Left and right flanking sequence of about 250bp long surrounding a SSR loci can be used for primer design. The tabular result section of SSR search contains a 'Details' link for each record, which opens window displaying the flanking sequences surrounding the corresponding locus. Clicking on Primer3 button it submits the loci and surrounding flanking sequence to Promer3Plus for primer designing. The reating region gets surounded by a pair of square brackets ([]) to exclude it from primer designing. Details about primer3Plus can be found here
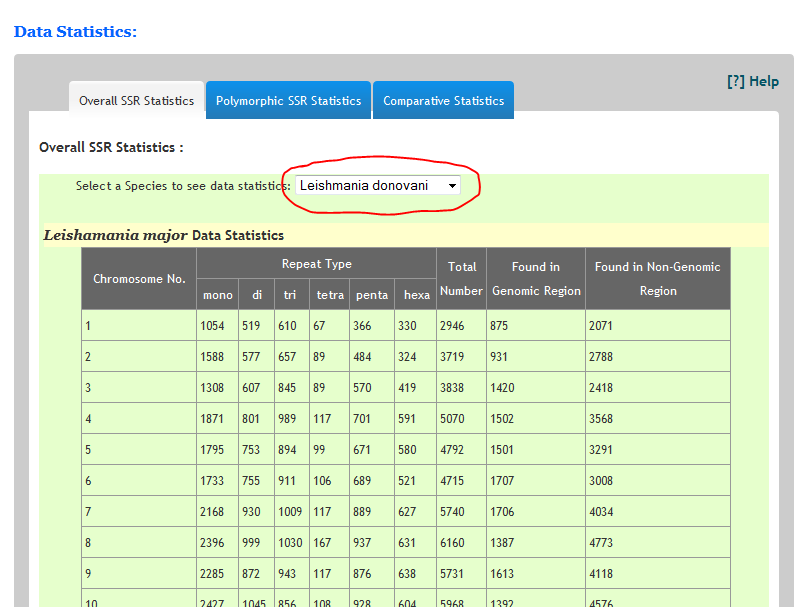
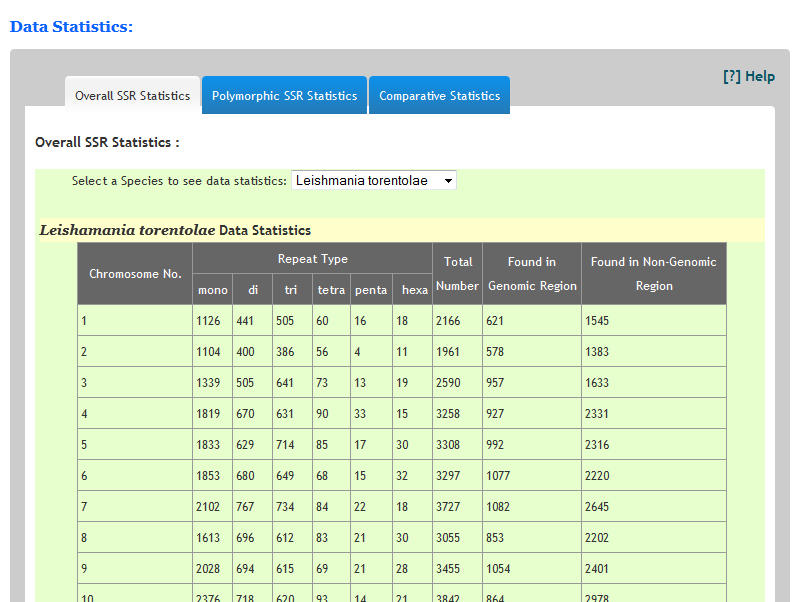
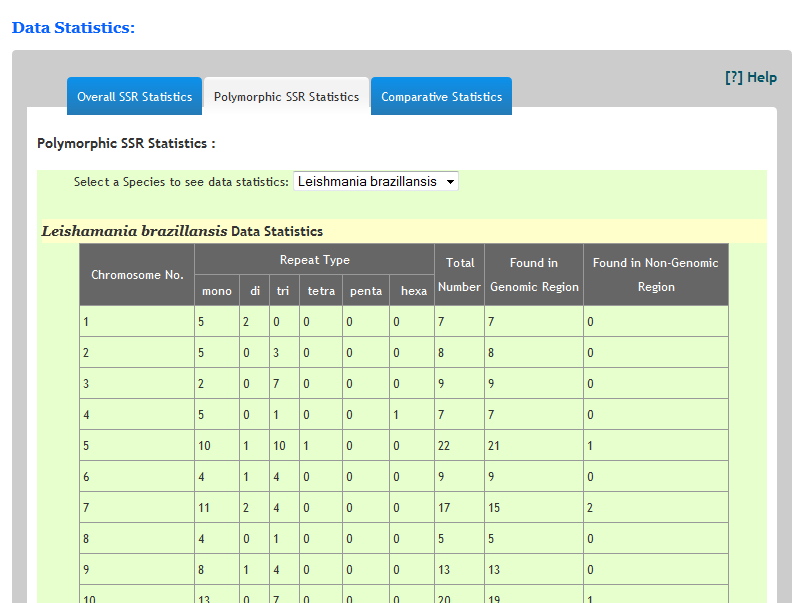
How many repeats are found in any species?The statistics of each species can be seen on Statistics page. Just navigate to statistics.html page. Select a specific organism from the list and you can see the chromosome wise distribution of repeats in the corresponding species.Similarly predicted polymorphic SSRs can also be seen under polymorphic Statistics tap.
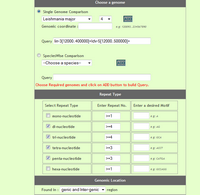
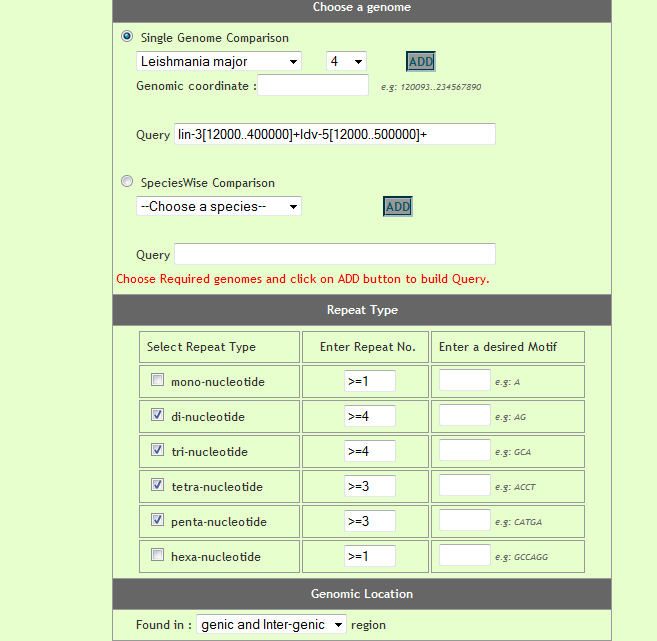
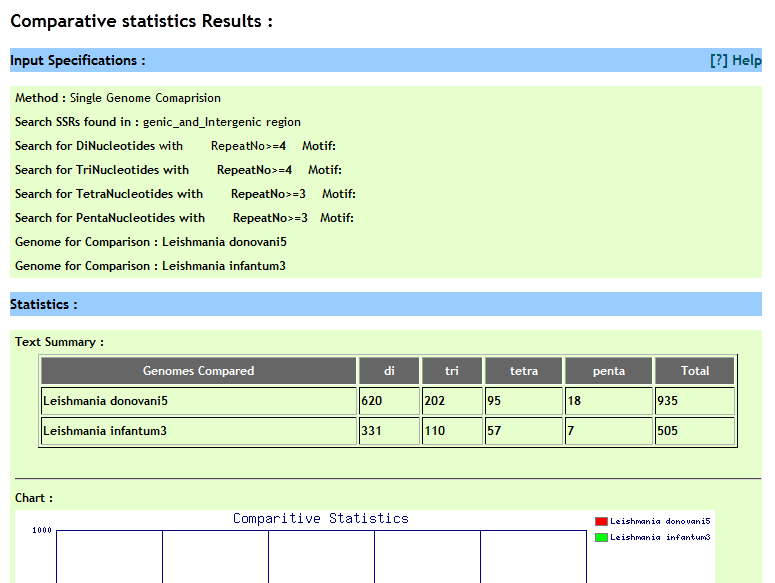
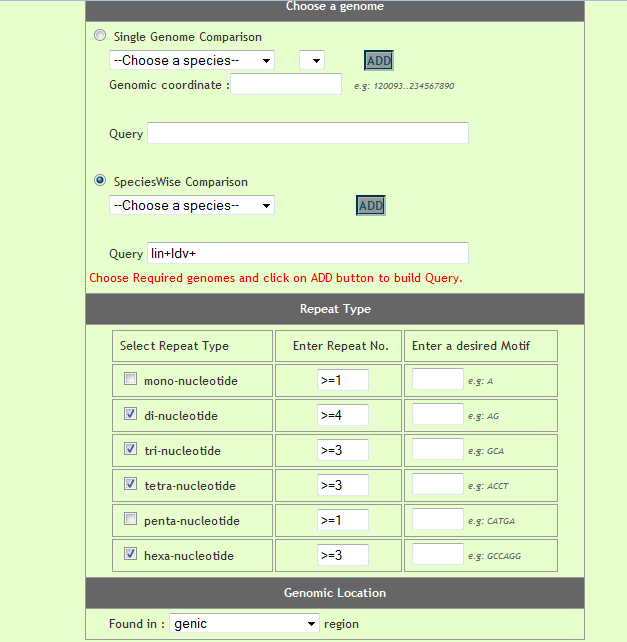
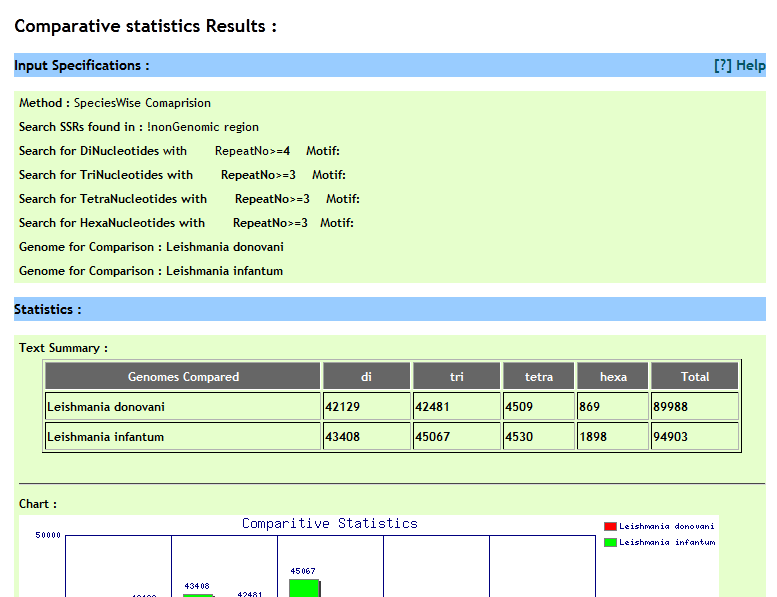
User can compare perfect repeat statistics in different desired species. He can either compare statistics from two(or more) chromosomes or can compare statistics of repeats from present on all chromosomes of the respetive species. Optionally user has options to speficy genome coordinates, repeat motif besides repeat type and repeat unit. For comparison user need to build a query by selecting different options and cliking on 'Add to query' button, iteratively.
How to extract SSRs in your own sequence?User can upload a single nucleotide sequence and extract the sequence repeats present in it. User can optionally upload a .ptt file to annotate the location of extracted repeats in genic and inter-genic regions. What is the database schema?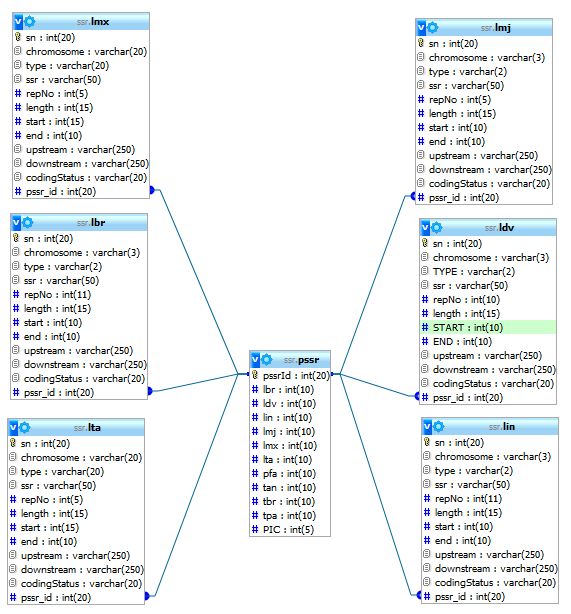
|
| Powered By Dept. of Bioinformatics, Rajendra Memorial Research Institute of Medical Science | FEEDBACK |